The Third Wave of Degrader Discovery
Proteins occupy a special place in the biological domain as the master regulators of biochemical processes in the cell. They accomplish this as enzymes, hormones, transporters, signal transducers, and other complex molecules to ensure the cell maintains a homeostatic balance. This system, while amazingly resilient, breaks down when those same proteins go awry, and it is the hallmark of most diseases, from cancer to neurodegenerative and autoimmune disorders. Current therapeutic interventions are designed to target these “rogue” proteins specifically, with cutting-edge technologies such as gene editing by CRISPR and mRNA gene therapies leading the way. CRISPR-Cas9 technology is an irreversible process. Safe and effective delivery systems that ensure little to no off-target effects are a significant area of concern, especially for neurological conditions like Alzheimer’s or Parkinson’s Disease.
Similarly, mRNA therapies face stability and immunogenicity issues that prevent their application to the clinic. Other traditional methods that use small molecules or peptides and act on the principles of inhibition make them incompetent to reach around 80% of the proteome.
So, what if drug discovery platforms did more than try to inhibit pathogenic proteins; what if there was a way to eradicate them?
Targeted Protein Degradation
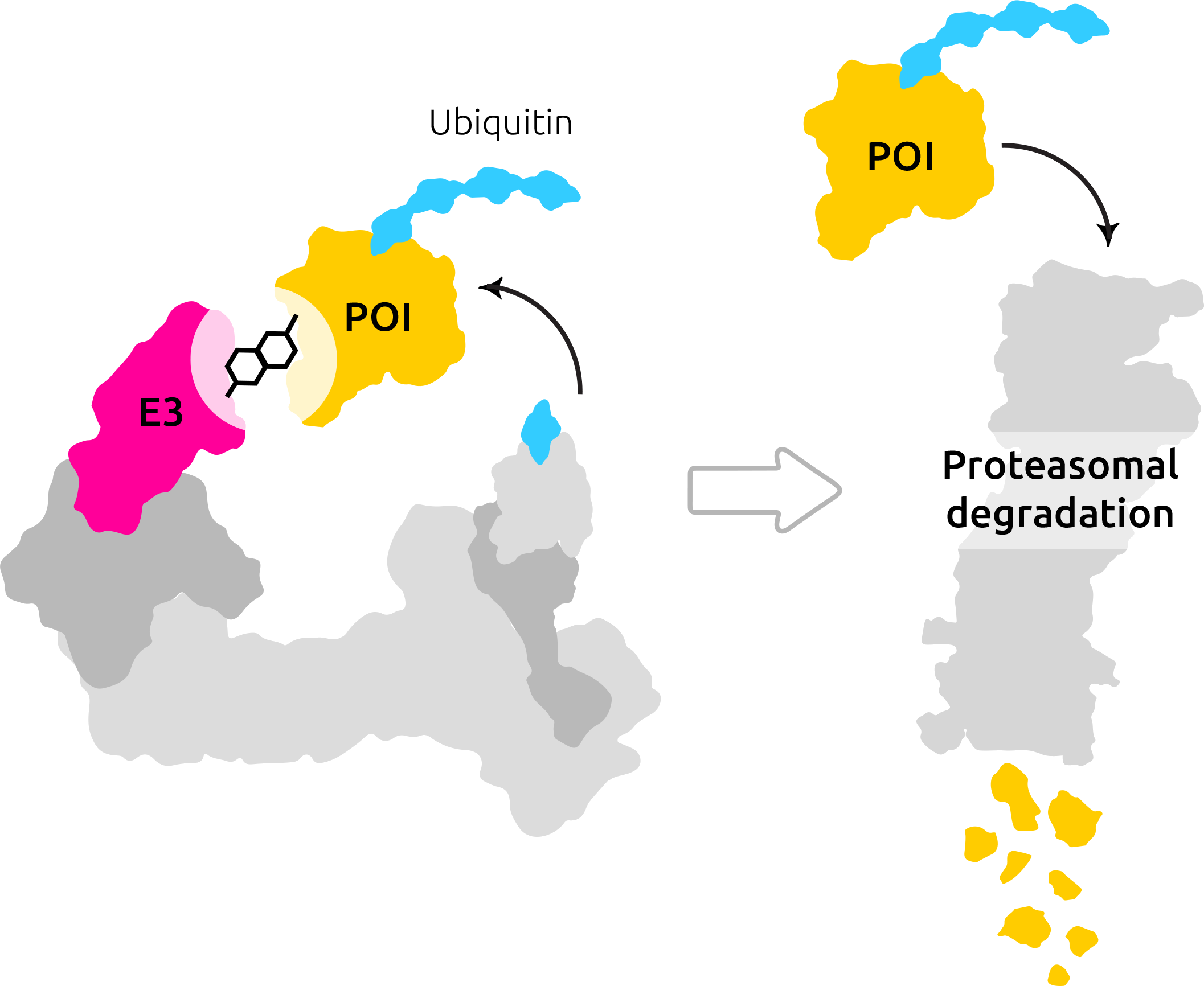
A niche and emerging player in small molecule drug discovery is TPD, which eliminates faulty or misfolded proteins in the cell. Molecular glues are small molecules that induce proximity between interacting proteins and stabilize protein-protein interactions. This property becomes helpful in the context of a particular class of molecules: the E3 ubiquitin ligases. An E3 ligase adds ubiquitin tags onto its substrate (target of interest) and marks it for proteasomal degradation. Molecular glues stabilize the bond between the ligase and the target, thereby strengthening their interaction. The ligase, target, and molecular glue thus form a ternary complex, a key species that drives the catalysis of ligase bond formation. In this way, the ubiquitin-proteasome system (UPS), a cellular system that tags misfolded or disease-causing proteins for degradation, might enable the degradation of the ~ 80% pathogenic proteins that are currently considered untreatable by modern drugs.
Now, why is TPD considered to be the new frontier in drug development?
If you think about it, there are apparent advantages such as:
- High selectivity due to the ternary nature of interactions between the degrader, target, and E3 ligase.
- Efficient target coverage due to the transient binding compared to occupancy-driven small molecule inhibitors.
- Obliteration of the target is opposed to inhibition.
- Low concentration levels of degraders in the application due to their nature of being recycled.
- And many more.
We want to address selectivity in particular in this blog entry.
It has become clear that tuning selectivity with degraders will ultimately depend on the available E3 ligases and their different cell expression profiles.
Molecular Glues versus Hetero-bifunctional Degraders
Similar in function but different in mechanism to molecular glues are the hetero-bifunctional degraders. Hetero-bifunctional degraders contain an E3 binding moiety that binds to the E3 ligase and a ligand or warhead that attaches to the target, connected via a linker. And although predictable concerning their binding to the target protein, their high molecular weight, and poor cell permeability impede their pharmaceutical application. In contrast, molecular glues were discovered serendipitously, and binding to the target protein in most cases is unknown. But where they lack in unpredictability, they more than make up for in “drug-likeness” as they are small molecules with a propensity to cross cell membranes. Both attractive properties make them suited for drug design.
The Waves of Degrader Discovery
The last couple of years have seen a growing interest in monofunctional and hetero-bifunctional degraders.
First Wave
Arvinas leads the pack as the first to do a clinical study of an androgen receptor degrader. Now it is joined by more than 15 degraders, set to come into the clinic by the end of 2021.
They are joined by C4 Therapeutics, and both companies contribute to the understanding in the field of degrader discovery with their drug development programs.
These companies were facing the problem that much of inhibitor optimization has been well-founded in guiding medicinal chemistry principles encapsulated in, e.g., Ro5.
As they were starting to go beyond the rule of five, it was expanding the scope and knowledge in medicinal chemistry.
Second Wave
The second wave of degrader discovery includes players like Amphista Therapeutics, whose platform operates independently of traditional E3 ligases, and BioTheryx that repurposes immunomodulatory IMiD compounds as oncology-based targets. Similarly, Kymera Therapeutics has focused on selectively developing its pipeline on cancers and immune-inflammatory diseases. The list of biotech companies that have broadened the druggable space by developing drugs with previously unknown binding sites continues to grow.
It is necessary to understand the difference between these two waves:
On the one hand, IMiDs were discovered serendipitously and used clinically without knowing their mechanism of action. Just through recent studies, science understood that they work as degraders, and therefore they gained industry interest.
On the other hand, hetero-bifunctional degraders came from a parallel line of investigation trying to design this modality artificially.
The problem with these hetero-bifunctional degraders is that we are inherently drawn to work on targets where there are already known ligands, which biases the discovery towards spaces where chemists have already worked. Whereas the IMiDs can degrade targets that weren’t considered to be targeted in general. To give a few examples: ZINC fingers, transcription factors, etc.
Another major drawback of the first wave hetero-bifunctional degraders is that they are large compounds with limited cellular bioavailability. Their cellular concentration can be 1000 or 100000x times lower than small molecules.
However, it is crucial to say that even though you might end up working on the known chemical matter with hetero-bifunctional degraders, it doesn’t necessarily mean that these molecules don’t have differentiated pharmacology.
Now, some of the critical aspects research and industry has become aware of after the second wave of degraders are:
a.) Identification of targets that are amenable for degradation
b.) Minimizing the compound size while trying to retain the pharmacological effects
c.) Translation from animal models to human studies
d.) Identification of the optimal dosages
e.) Expansion of the E3 ligase space for targeted protein degradation
Third Wave
As we have seen that some limitations became apparent during the first two waves of degrader design, such as bioavailability, the scope of TPD beyond cells and tissues, resistance and drug-like properties, including difficulty in routine compatibility with oral dosing regimens, the question remains:
How can we draw logical conclusions here and translate this knowledge into the most efficient research possible?
Based on these challenges in early-stage degrader discovery, it becomes clear that we need to predict properties associated with these molecules with certainty, as they go beyond the rule of 5. Implementing AI for data-centric drug discovery is essential to enable and accelerate critical findings related to binding affinity and pharmacological properties. Companies investing in these molecules need to understand fundamental questions related to bioavailability, mechanism of action, and whether or not the molecule can cross the blood-brain barrier in the case of CNS disorders. AI-driven applications can address and expedite some of these challenges.
Celeris Therapeutics is leading the way of the third wave of degrader discovery via their machine learning-enabled platform.
The CelerisTx perspective Part 1
Inducing TPD with ML-enabled interaction predictions
One remaining challenge includes understanding the binding behavior of all the molecules involved in TPD and their formation into a ternary complex.
Celeris Therapeutics determines the strength of the interaction between molecules in an automated manner, based on the 3D information of the surfaces of proteins and ligands and their physicochemical features (proteome-wide screening).
These automation steps include:
- Scan of protein surfaces (target and ligase) and identification of protein-protein-interaction scores (indicating weak/strong binding)
- Introduction of new chemical entities (NCEs) and ML-based screening against the target (precise specification of the positions for interaction and binding)
- Modification of the target surface after binding to the NCE (dynamics).
- Running interaction predictions between the modified target and ligase again and calculating a new PPI score.
- Calculation of the delta between steps 2 and 4 in the PPI score. If this delta is high, it shows that introducing the NCE has increased the interaction between the target and ligase significantly. Hence it predicts in an in-silico manner whether the NCE can glue these proteins together.
Stay tuned for the next parts of the Third Wave of Degrader Discovery!