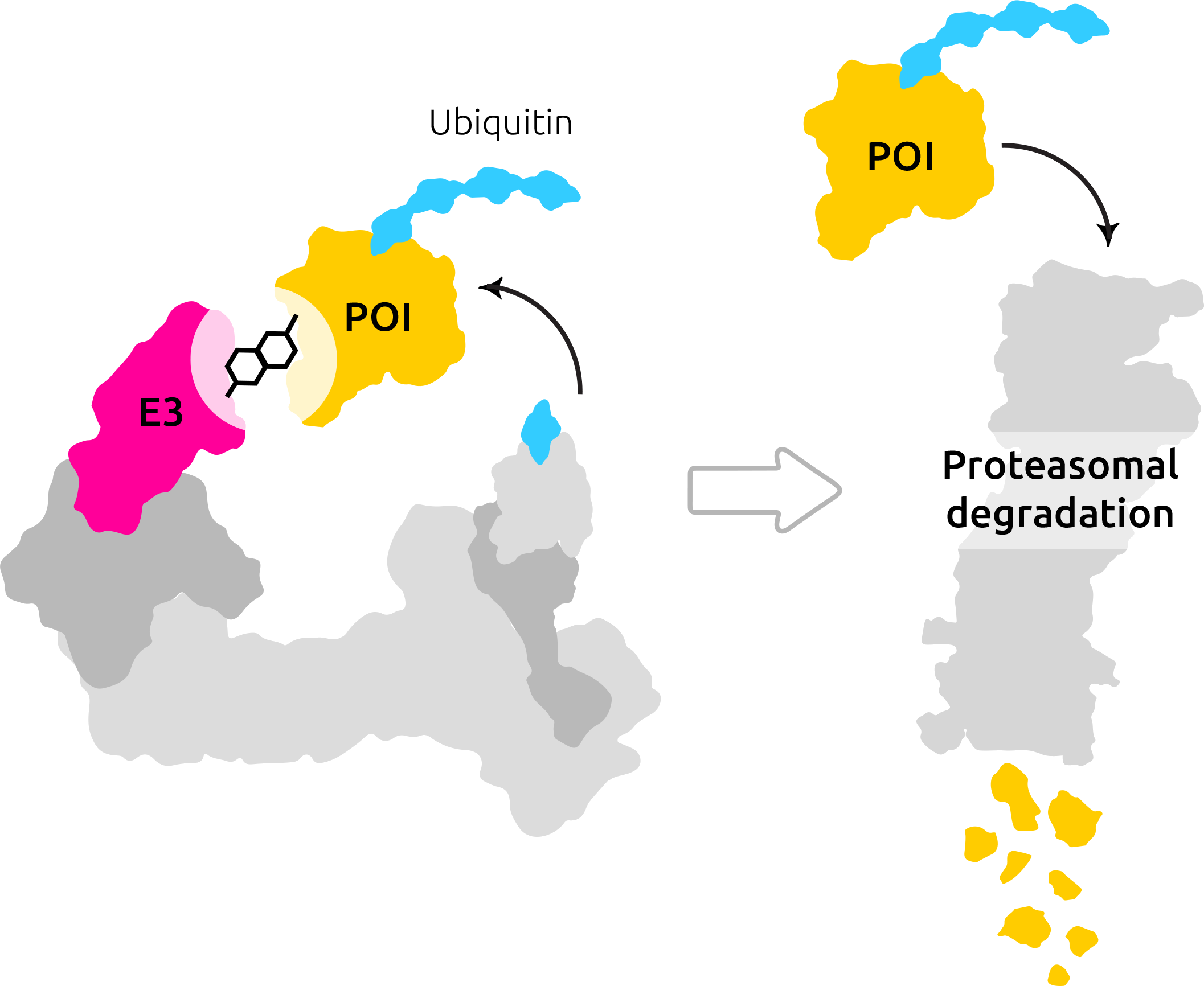
With the new book The Science and Technology of Growing Young by Sergey Young, we, Celeris Therapeutics, an AI company in this space, had to analyze the book through our AI lense and pose potential approaches to tackle the problems outlined in the book.
“Aging, quite simply, is a loss of information”, Dr. David Sinclair
If one peels through the layers of aging problems and rephrase these challenges in terms of information and data, one can see that aging can be tackled computationally with AI.
The book has two main directions – The Near Horizon of Longevity and The Far Horizon of Longevity. We will focus on the former, with brief input to the latter at the end.
The Near Horizon of Longevity
One of the root causes of understanding complex systems is the fallacy of the correlation implies causation.
Let’s take one of the prominent longevity theories, telomere shortening. The short version is that the telomeres, repetitive nucleotide sequences associated with proteins that lie on both ends of our chromosomes, directly impact longevity – to be precise, the length of telomeres. Scientists observed that shorter-telomere mice lived shorter and vice versa. We rushed to confirm this theory to humans, and ambiguity happened: Even though there were promises, excess telomerase was also linked with cancer. So which one is it, is the telomeres length cause of longevity, or is the length of the telomeres the effect of longevity. We need more data to prove it!
Sergey Young outlined in the third chapter four leading breakthroughs that will have a significant impact on longevity. Here is our (AI) take on them.
- The genetic engineering breakthrough.
The Human Genome Project was finished in 2003, and it successfully sequenced the entire human genome. Meaning we had the information of the whole twenty-five thousand individual genes. Gene sequence allows us to predict many hereditary diseases, the probability of getting cancer, and many other unknowns where we still did not determine the cause and effect relationship. You may be wondering now, given that our genes do not change much from birth, what are other implications of gene sequencing. As Sergey Young points out, our epigenome, the system of chemical modifications around our genes that determines how our genes are expressed, does.Furthermore, according to longevity scientists, the biological age of your epigenome, not your actual age, seems far more critical. Why is this important for ML? Horvath clocks, biological age measuring solutions enable measuring of “aging score” and effectively measuring the chemical activity within the epigenome. All this sums up that now we simulate longevity experiments and observe the effects in a human cell long before this human dies. Deep learning can be used to improve the Horvath clock accuracy.Another enabler is the price. The price to perform genome sequencing has come down to a couple of hours and hundreds of dollars. The problem (opportunity) is that the size of the data is enormous. One genome contains three billion nucleotide base pairs. The chance to analyze this and make a subsequent inference is considerable. Change just one letter (one nucleotide) in the genome, and one can cause severe side effects. One concrete example of understanding our genome and the effect genes cause is the so-called gene therapy. It works by effectively focussing on genetic modifications of cells to produce a therapeutic effect. You may have heard of the CAR T-cell therapy. A cancer treatment therapy based on gene therapy where patients own immune system T cells are modified to fight specific types of cancer. Machine learning can be used to improve the manufacturing process of CAR T-cells.
- The regenerative medicine breakthrough.
Aging wears down the bodily systems and functions. Achieving longevity does not only mean counting up the years but making the years count. For that, we need technologies that can repair these functions. One of those technologies is stem cell therapies. It has been talked about for years, but what has been emerging and further enhancing this subfield is, you guessed it, AI. Ways AI can aid multitude, with a formal overview of tackling different parts of stem cell technology with AI available here.
- The health-care hardware breakthrough.
Out of all potential breakthroughs, this one holds the most sway. Yearly, out of the 60 million lives lost, we can contribute more than half of that to reversible diseases if caught early. The problem is if we are not monitoring frequently and granularly enough to react in time. We are posing our current approaches as “reactive” and not “proactive.” If we can get this data early and accurately enough, we will diagnose the condition at hand. This brings us to one of the essential principles in AI. It’s the data that’s the differentiator, not the machine learning algorithms. If you can gather the quality (labeled) data, the algorithms operating upon this data are not that important. And yes, current wearables are very constrained in function. But this is changing fast, and look at this recent Nature publication. We can measure deep tissue processes without invasive surgery. He who owns the data owns the AI.
- The health data intelligence breakthrough.
Sergey Young argues that justifiably the seismic shift underpinning the Longevity Revolution is the amount of and quality of data coming in and from these devices. This is also a massive enabler for Celeris, allowing us to target the undruggable proteins and learn what makes a ternary complex part of the degradation process. More on that you can find in this blog post. But another vital aspect that Sergey mentions is the practice of precision medicine. It involves customizing health diagnostics to individual characteristics. Here is the problem: Privacy issues. For the machine learning algorithms to operate on the data, one needs to access them first. Sending this data to a server where the diagnostic computation will be performed is a problem.
There are two straightforward solutions: on-edge machine learning and differential privacy. The first aspect leverages the fact that our current devices (not only phones) have become exceedingly powerful. They can perform a computation reserved for personal computers or computing clusters just a couple of years ago. The computer used to send the first man to the moon is now weaker than the current phones. This implies that the expensive machine learning calculations can be offloaded to phones and smaller medical devices – obviously, not all components, but just the ones that need the personalized data. For example, one company can train the model offline, meaning on the servers, and then send the model to the patient’s devices to make the inference on the device. Avoiding the privacy problems as a whole. The second paradigm is that of differential privacy. The gist of it is that these are the standard machine learning algorithms with carefully “injected” noise, which mask the data and mathematically guarantee privacy. The data will never be seen, and the algorithms can still extract patterns (make the diagnosis) on this noisy data. This is nothing new; these kinds of algorithms are precisely what Google and Apple use when they process our private data. Or at least they claim they use.
NOTE: Privacy discussion and its solutions will be even more critical as we move towards the internet of bodies (IoB). All of the sensors that measure our data are paired between themselves and other people’s data. Since collective intelligence is more significant than single, this step will be essential to get better AI. But managing it will introduce security challenges on its own. Two solutions of which we proposed above.
All this abundance of data is intertwined with better diagnostics. Sergey Young outlined novel, non-invasive, cheap, and highly effective diagnostic methods:
- Liquid biopsy. Enables examination of bodily fluids like urine, saliva, spinal fluid, or blood to find traces of diseases, examine DNA or RNA. All while we are non-invasive and cheap. The fact that we can extract a lot of information from a single drop of blood opens many doors for diagnosis. As we understand and further analyze, e.g., blood data, we can make better machine learning algorithms. Some companies are already doing this, where they have built in a sense a diagnostic recommender system based on your personal blood information. Data? liqDB is a small-RNAseq knowledge discovery database for liquid biopsy studies. What could you do with it? With this and plenty of other liquid biopsy databases, labels indicate different diseases based purely on bodily fluids measurements. There is an opportunity to expand this to other, not labeled diseases and see if accurate inference can be performed.
- Genetic diagnostics. In the genetic engineering breakthrough, I defined one potential improvement of the Horvath clocks. Still, the reality is companies and research are abundant trying to make actionable insights from the genome. One exciting application is customizing your diet to your genetic makeup. The real challenge, in this case, is not the ML algorithms but creating the data to help make a correct inference. Data and ML Application? As mentioned in the 1. The genetic engineering breakthrough. But the applications and databases are vast. A comprehensive summary can be found here.
- Epigenome diagnostics. As Sergey summarizes, epigenome may prove more critical in early disease diagnosis than the genome. Epigenetic changes, changes in the chemical compounds and processes that regulate gene expression, can be used to find diseases much earlier than the genetic approach. And are far more accurate in predicting these diseases. Data? Epigenetic Tools What could you do with it? From supervised to unsupervised to RL, the extent of possible application is vast. An interesting example of unsupervised learning was done here. They profiled a high-resolution comparative genomic hybridization (aCGH) and RNA sequencing (RNA-seq) to analyze chromosomal alterations and dysregulated gene expression in tumor specimens of patients with fibrolamellar hepatocellular carcinoma. It is a mouthful, just showcasing that these types of AI applications can not be done without a multidisciplinary approach.
- Microbiome diagnostics. The fact is, the digestive tract hosts more microorganisms than the number of cells in our entire body. But the more interesting fact is that the combination and performing actions of these microorganisms are different for every individual. And these unique combinations, it turns out, are a good indicator of certain diseases. The microbiome is very closely linked to health and longevity. The problem is how do we crunch the data? Again blindly applying the ML on whatever is available out there in regards to microbiome will not work. True enablers here will curate datasets, paired with domain knowledge of clinical experts and only then applying AI to get the superior inference. Data? HMP Database human microbiome project. What could you do with it? An extensive review of methods can be found here. They are taking just one example of applying deep learning, DeepARG networks trained to predict antibiotic resistance genes (ARGs) from metagenomic data.
In our previous blog, we talked about nine leading “causes” of our decline, the nine equestrians of an internal apocalypse. Sergey outlines the tenth one as:
Protein crosslinking: This is a phenomenon where multiple separate proteins become bonded together by a sugar molecule in a process called glycation. Depending upon where this occurs, it tends to correlate with different signs of aging — wrinkles, arteriosclerosis, cataracts, and kidney failure, to name a few.
Data? ProXL (Protein Cross-Linking Database): A Platform for Analysis, Visualization, and Sharing Protein Cross-Linking Mass Spectrometry Data. What could you do with it? Developing an ML to identify where this bondage/linkage occurs and what kind of damage it will cause. If labeled data is hard to obtain, clustering techniques can be used to analyze already known bonding regions and try to abstract from there.
The Far Horizon of Longevity
Two main enablers outlined here were quantum computing and artificial general intelligence. We did not want to analyze this part to a greater extent because there is a high probability we would speculate. Technological advances are only apparent in hindsight. And AI had its winters. We will instead try to dissect the current state of this technology and what can be applied.
Even though there is a lot of hype around the execution speed that quantum computing brings, this is not (currently) true for all of the computational problems. Depending on the steps that need to be executed, quantum computing might be slower than the traditional computing paradigm. Take, for example, quantum machine learning. Google developed a method to evaluate the suitability of applying quantum machine learning given the data at hand. And the conclusion is following:
Moreover, a key takeaway from these results is that although we showed datasets where a quantum computer has an advantage, classical learning methods were still the best approach for many quantum problems.
Currently, there are companies like AWS offering quantum computing services like Braket, and given the fact, there is an ample space of molecules to be parsed, computational needs are enormous. Hence there is no surprise that there is research in applying quantum machine learning to drug discovery space. An opportunity to use this to our degrader approach is also available. The maturity is still coming of age, and there is a lot of potential in classical computing to be exploited. Take, for example, the active learning domain of ML on our ternary complex predictions. Here one can parse huge data spaces (molecules) without relying on brute force computation but intelligent heuristics.
To talk about the state of artificial general intelligence (AGI), we first have to define it. Taking it from Sergey’s book, AGI “should devise a solution to any task through observation, research, and application of past experiences.” While there are different approaches to achieving AGI, and even reputable sources claiming this will not happen, the current consensus to at least get closer to AGI is applying reinforcement learning. Even a DeepMind publication is claiming this is the case, called “Reward is enough” since the basic premise of this ML domain is training an agent to perform particular action purely by observing the environment and then incentivizing him to perform desirable action rewarding him. When looking at the current development of this discipline, the status is conflicted.

From https://www.alexirpan.com/2018/02/14/rl-hard.html
While there is a lot of research being done, the number of real-world applications is limited. Reasons are multitude, and it is extensively summarized in this article. This is not to say that it can not be done, and there are already people applying this in the drug discovery space, but the scalability and the production level maturity will be a question.