Proximity-inducing compounds
Celeris Therapeutics is the pioneer in AI-driven proximity-inducing compound (PIC™) design focused on targeted protein degradation. We translate 3D structural data into much-needed medicines using AI and induced-proximity.
What are PICs™?
PIC™ drugs represent a new class of therapeutic modalities able to hijack naturally occurring biological machines.
Processes such as autophagy, folding via chaperones, lysosomal and ubiquitin-mediated proteolysis play an important role in protein regulation. PICs™ allow us to take advantage of these processes and hence provide an unprecedented potential to further expand therapeutic horizons.
Targeted Protein Degradation
Mono- and bivalent degraders are shifting drug development paradigms from a focus on inhibition towards the removal of pathogenic proteins.
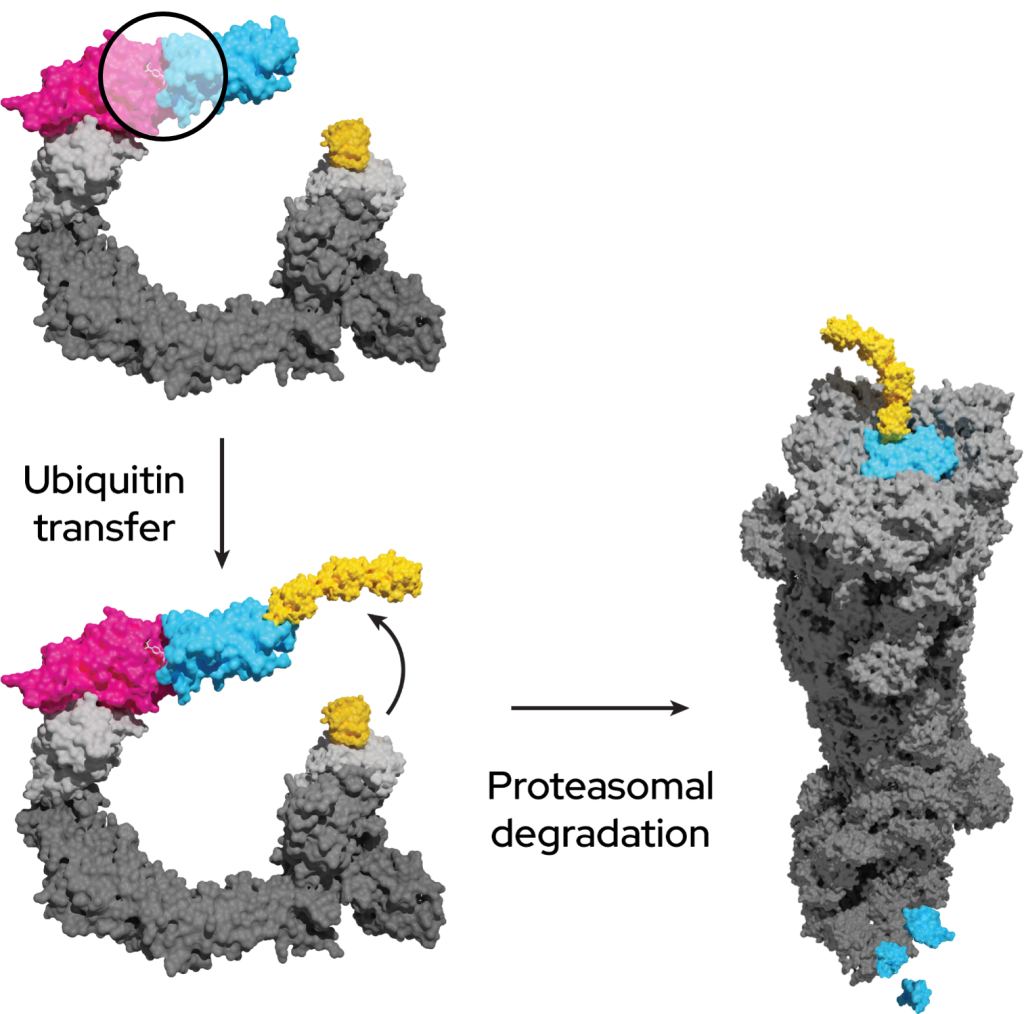
A key focus of targeted protein degradation is the development of proximity-inducing compounds that move a pathogenic protein into cellular machinery, such as lysosomes, chaperones or the ubiquitin-proteasome system (UPS).
Our wholly owned drug development pipeline hijacks the UPS via PICs™.
The UPS is present in all eukaryotic cells and selectively removes proteins in three steps:
- The ubiquitin-activating enzyme (E1) activates ubiquitin, which is then transferred to a ubiquitin-conjugating enzyme (E2).
- Ubiquitin on the E2 enzyme is conjugated to lysine residues within the protein of interest, or the N-terminal amino group, by a ubiquitin (E3) ligase. E3 ligases can be highly selective due to protein-protein interactions between the E3 and the protein of interest.
- Additional ubiquitin molecules are conjugated onto the first, forming a polyubiquitinated adduct recognized by the proteasome leading to degradation of the protein of interest.
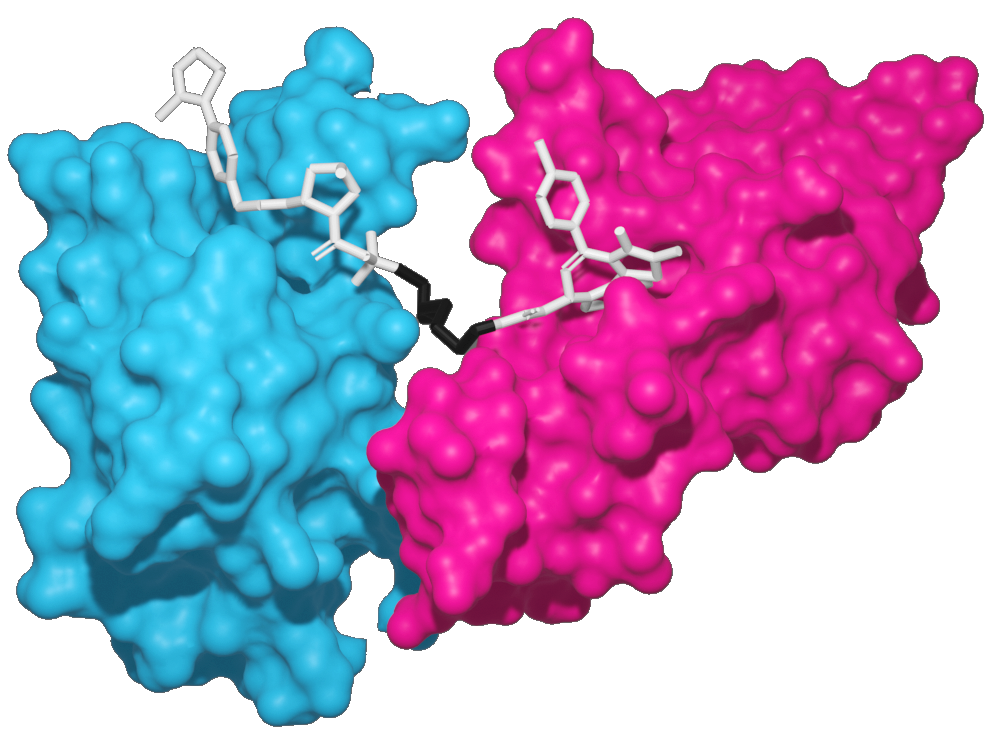
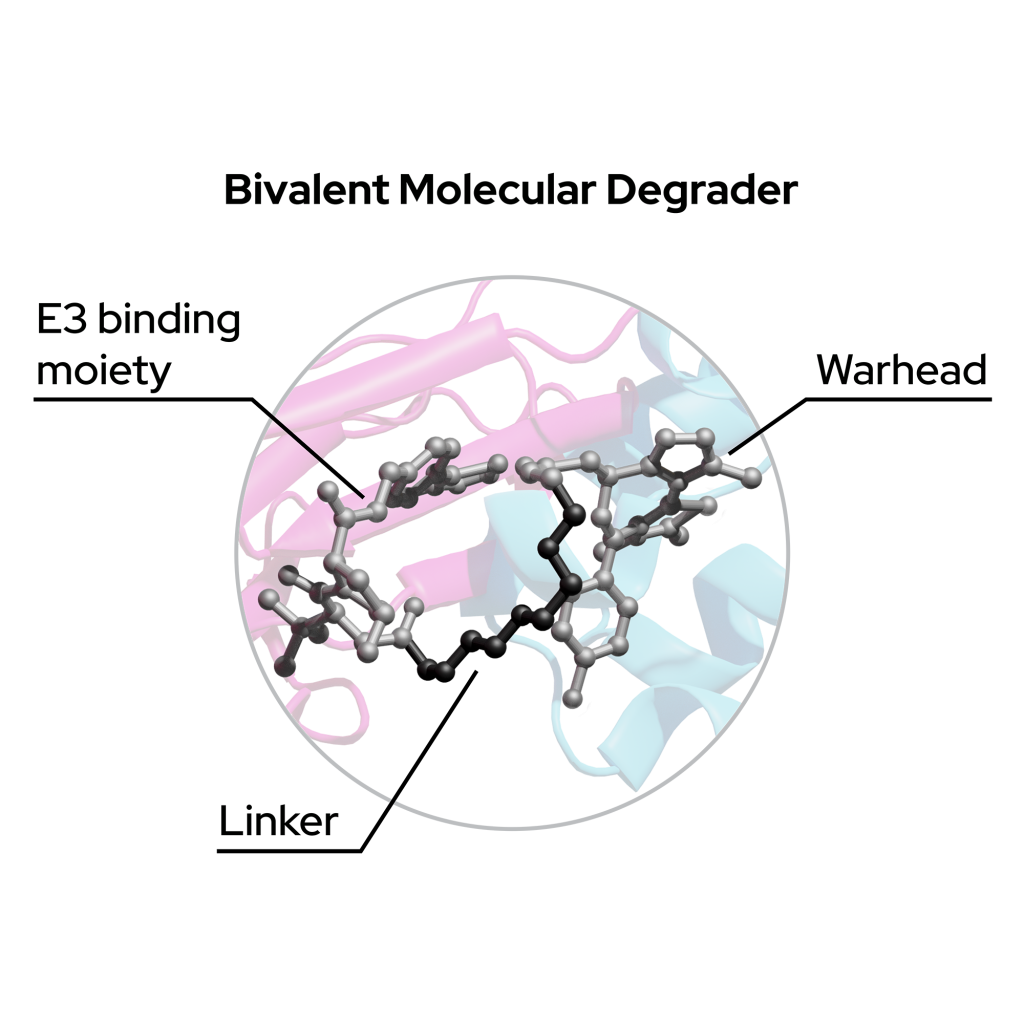
Bivalent degraders
Bivalent degraders consist of three compound regions:
- POI target ligand – Warhead that binds to the Protein-of-Interest
- Molecular machinery ligand – Binds to, e.g., lysosome, E3 ligase
- Linker – A region that links these two ligands
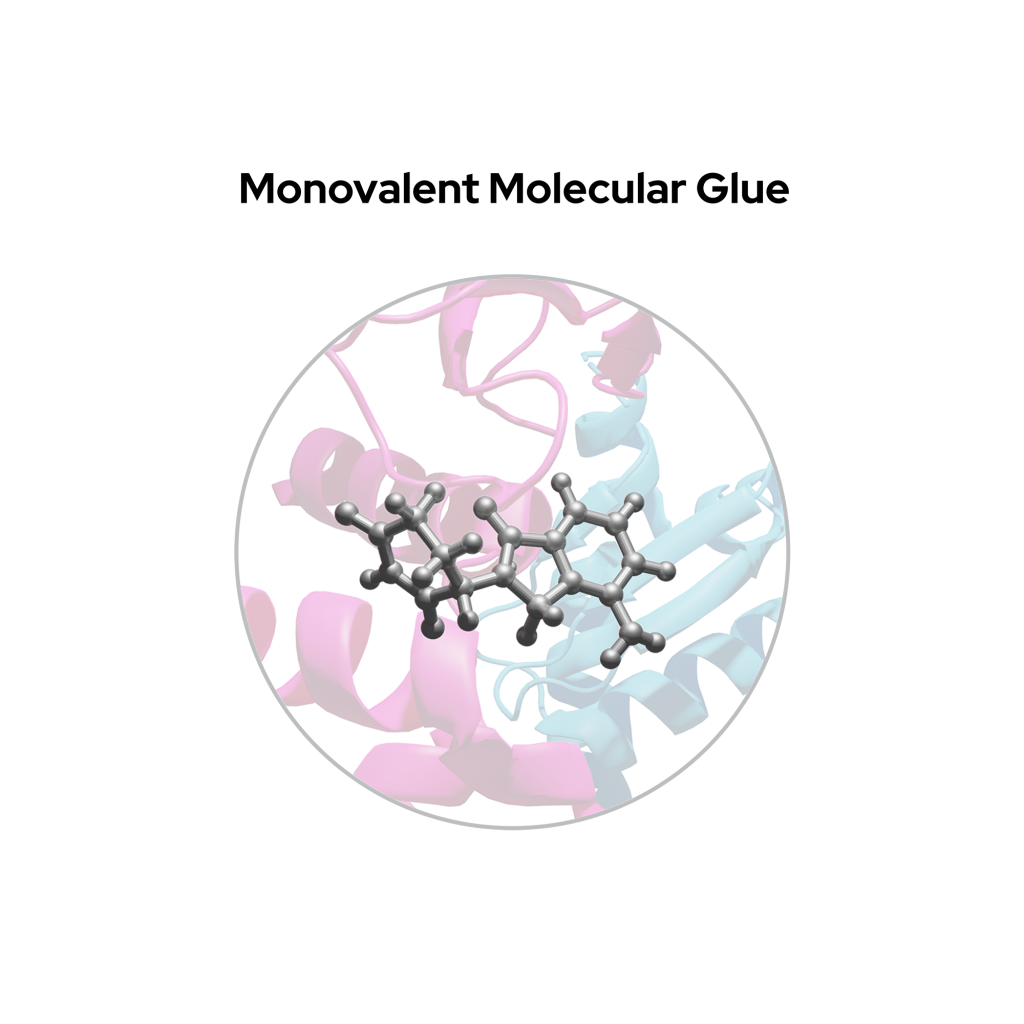
Monovalent degraders
Monovalent degraders, often referred to as molecular glues, consist of only a single compound region that glues an E3 ligase to the POI by altering the surface of an E3 ligase substrate receptor, inducing a naturally occurring protein-protein interaction interface.
Advantages
Target Intractable Proteins
Traditional drugs try to inhibit the functions of pathogenic proteins, but only 10-20% of all pathogenic proteins can be inhibited. PICs™ show promise in overcoming yet-to-be-drugged targets connected to diseases with high unmet medical needs, such as Parkinson’s. Many ongoing clinical studies back these claims. 1, 2, 3, 4, 5
High Target via Protein-Protein Interactions
Due to their characteristic of stabilizing cellular machinery and target protein interactions, PICs™ exhibit exceptional selectivity compared to small molecule inhibitors. 8, 9, 10
Ease of
Manufacturing
PICs™ are chemically-derived small molecules and not extracted from living organisms. As a result, chemical synthesis can be scaled efficiently once the synthesis pathway is identified.
Reduction of Off-target Effects
Cellular machinery is differentially expressed in various tissues, cells and compartments. Applicability to specific tissues and cell types reduces the off-target effects of PICs™. 7, 12, 13
Disrupting Scaffolding Functions
Although small molecule inhibitors silence distinct domains of proteins, scaffolding functions of targeted proteins outside the small-molecule binding site can remain active. By regulating the overall protein concentration, PIC™ degraders are more likely to recapitulate the phenotypic data seen in knockout (CRISPR) and knockdown (RNAi) target validation assays.
Oral
Bioavailability
Therapeutic strategies which modulate gene function by blocking gene expression might have a similar impact on phenotypes compared to a PIC™ strategy. However, knockout (CRISPR) and knockdown (RNAi) therapies have significant administration limitations, whereas PICs™ provide the unique pharmacodynamic advantages of small molecules – including oral bioavailability.
Potency Disconnected from Affinity
Inhibitors rely on the occupancy of a binding site, which requires high-affinity or high concentration to compete with endogenous substrates. Due to their event-driven pharmacology, PICs™ are catalytic in nature and consequently show higher potency. 12, 13, 14
Clinical Validation
While other PIC™ modalities are being explored, Celeris Therapeutics’ pipeline is centered around PICs™ that initiate targeted protein degradation. Degraders have been clinically validated with the first bivalent degrader drugs entering clinical trials in 2019 and promising interim Phase 2 data reported in 2020.
-
Kathleen M. Sakamoto, Kyung B. Kim, Akiko Kumagai, Frank Mercurio, et al., (2001). Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. PNAS 15, 8554-8559
-
Asher Mullard (2021). Targeted protein degraders crowd into the clinic. News, Nature Reviews Drug Discovery 20, 247-250.
-
Eric S. Wang, Alyssa L. Verano, Radosław P. Nowak, et al., (2021).
Acute pharmacological degradation of Helios destabilizes regulatory T cells Nature Chemical Biology. 17, 711–717 -
Schneider, M., Radoux, C.J., Hercules, A. et al. The PROTACtable genome. Nat Rev Drug Discov (2021). https://doi.org/10.1038/s41573-021-00245-x
-
Pujols J, Peña-Díaz S, Lázaro DF, Peccati F, Pinheiro F, et al., (2018). Small molecule inhibits α-synuclein aggregation, disrupts amyloid fibrils, and prevents degeneration of dopaminergic neurons. PNAS 41, 10481-10486
-
Piazza I, Beaton N, Bruderer R, Knobloch T, Barbisan C, Chandat L, et al., (2020). A machine learning-based chemoproteomic approach to identify drug targets and binding sites in complex proteomes. Nature communications 11, 4200. doi:10.1038/s41467
-
Kostic, M., & Jones, L. H. (2020). Critical Assessment of Targeted Protein Degradation as a Research Tool and Pharmacological Modality. Trends in pharmacological sciences, 41(5), 305–317.
-
Smith, B.E., Wang, S.L., Jaime-Figueroa, S. et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat Commun 10, 131 (2019).
-
Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, Wang J, Hamman BD, Ishchenko A, Crews CM. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem Biol. 2018 Jan 18;25(1):78-87.e5. doi: 10.1016/j.chembiol.2017.09.010. Epub 2017 Nov 9. PMID: 29129718; PMCID: PMC5777153.
-
Philipp M. Cromm, Craig M. Crews,Targeted Protein Degradation: from Chemical Biology to Drug Discovery (2017), Cell Chemical Biology, (24)1181-1190,
-
Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16(2):101-114. doi:10.1038/nrd.2016.211
-
Guo, WH., Qi, X., Yu, X. et al. Enhancing intracellular accumulation and target engagement of PROTACs with reversible covalent chemistry. Nat Commun 11, 4268 (2020).
-
Proof-of-Concept with PROTACs in Prostate Cancer (2020) Cancer Discov. (10) (8) 1084
-
Katherine A. Donovan, Fleur M. Ferguson, Jonathan W. Bushman et al., (2020). Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell. 183, 1714-1731