In this podcast Oskar Hoff, Head of Medicinal Chemistry, talks to Hosein Fooladi, CSO, and Noah Weber, CTO, about state of the art AI technologies and how it connects to Celeris Therapeutics and what they foresee for the next ten years.
Insight One
- The beginning of Celeris Therapeutics
- What role does AI play?
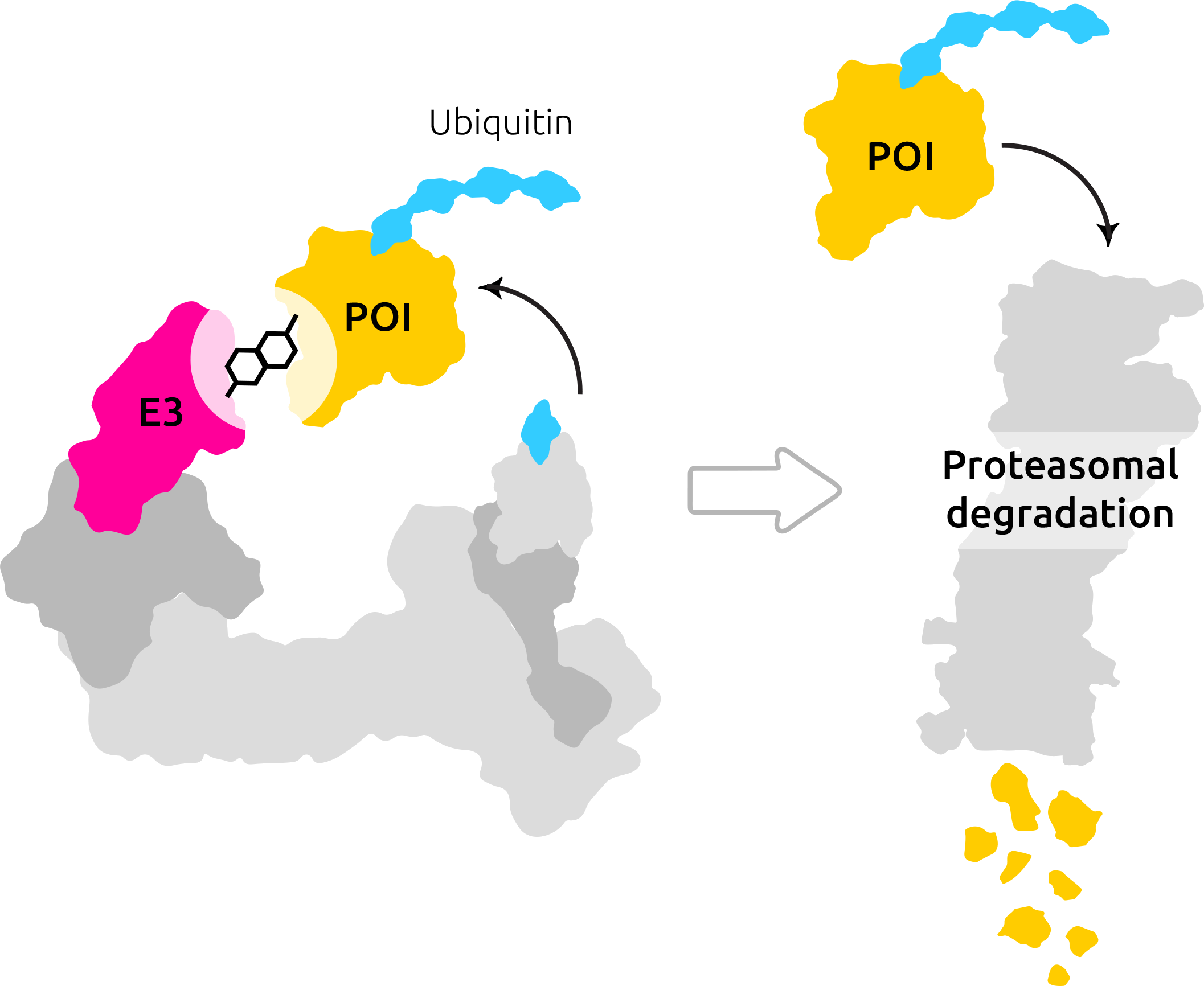
- What are degraders?
- Why degraders, and what’s the impact?
- How does a job as Machine Learning Researcher look like?
- How does a job as Computational Chemist look like?
- What’s Celeris One, and how does it work?
Transcript
Oskar: Welcome to the second episode of Insight One from Celeris Therapeutics, a deep tech startup in the intersection of machine learning and life sciences. My Name is Oskar Hoff, and I’m the Head of Medicinal Chemistry at Celeris Therapeutics and will guide you through this episode.
This series contains seven episodes, is called Insight One, and shows you the insights of Celeris Therapeutics.
Today I have Hosein Fooladi, the CSO, and Noah Weber, the CTO of Celeris Therapeutics, as my guests, and we’ll talk about the role that AI plays in our company.
Hosein, you were honored by the Iranian Ministry of Science and Research as an exceptional talent after being ranked in the top 0.1% in the national licensing exams. You have worked at the Cambridge Systems Biology Center as Machine Learning Researcher in Drug Discovery before joining a Cambridge-based in silico drug discovery startup. In 2021 you then joined Celeris Therapeutics.
After that respective and deep journey, I’m keen on getting an answer to a simple question: What does Celeris Therapeutics do differently?
Hosein: Thank you for the introduction, Amir. Indeed, I have much experience in AI-driven drug discovery, but that mainly shows me how much open space there still is. CelerisTx does a few things differently.
It’s the modality of the drugs. Only 10-20 % of all pathogenic proteins in the human proteome are considered to be druggable. The other 80-90 % is therefore called undruggable and is related to incurable diseases, such as Alzheimer’s and various types of cancers.
Instead of inhibiting functional sites of proteins, we exploit natural cell-based mechanisms to degrade proteins. That gives us the potential to degrade pathogenic proteins that are related to currently incurable diseases.
The second big differentiator is working on graphs and 3-dimensional representations of small molecules, proteins, and other macromolecules. The flourishing of graph neural networks have begun from 2-3 years ago, but in recent years, we are observing the power of these networks on real-world applications. We are leveraging their power for predicting molecular properties, the interactions between different molecules, and so on.
Also, we are working in the domain (targeted protein degradation) with very little/sparse available data. As a result, it is impossible to use big giant networks and traditional supervised learning methods for prediction. We should leverage the information from very little data, and this is where we are trying to apply active learning and the Bayesian optimization framework. We have the intention to entangle active learning with our wet-lab experiment and establish a hybrid approach that uses the full potential of machine learning with wet-lab data.
Oskar: Thank you very much, Hosein, for that comprehensive overview. Coming to you, Noah. You graduated in applied mathematics from the Technical University of Vienna and became one of only a few Kaggle grandmasters, a global competition platform for data science competitions, and already have much experience in ML engineering, research, and ML operations. From your perspective, what do you see as the main differentiator to other existing solutions, Noah?
Noah: One of the key differentiating points of Celeris is that it managed to answer Peter Thiel’s question, “what heretical view do you have” targeted protein degradation is nothing that other InSilico drug discovery companies are even considering doing. This is strange given that the next Nobel prizes are very likely going to go into the hands of initial inventors of this biotechnology. Automating this with ML is logical. But hindsight is 20/20. Pair this with our research, and you get Celeris.
Oskar: Thanks Noah for giving us that insight. What do you think, Hosein, what technologies might be worth watching in the upcoming 10 years when it comes to AI?
Hosein: it is hard to extrapolate for 10 years, but I will just provide my intuition. Two approaches are currently trendy: One approach reduces inductive bias and feeds the vast amount of data to a high-capacity network (transformer is a prevalent choice these days for that high-capacity network). The rationale is the by seeing the huge amount of data, the network will learn the right inductive bias. Also, I am considering AutoML in this category, in which we try to even learn the structure/building blocks of the network itself.
On the other hand, some people are arguing against this approach. They argue that even you feed tons of data to the network, you cannot answer “why” questions, and fundamentally for answering why questions, you need something beyond data. We hear the term “causality” and “causal machine learning” frequently these days, and the goal is to enable the system to answer questions related to interventions (in a world/distribution that is different from the current world/distribution). Answering these questions requires having information about the underlying causal structure.
So, I am more or less towards a hybrid approach that leverages the power of both sides. I think these are the approaches that are going to shine in the last couple of years.
Oskar: Thanks, Hosein. What do you think, Noah, given the current breakthroughs in biotechnology and AI, such as AlphaFold, what are the lessons learned from them?
Noah: Taking an example of AlphaFold, one of the major lessons is the marginal benefits of hard coding the domain knowledge. One of the DeepMind researchers’ approaches is that they did not perform a lot of feature engineering but let the system learn the problem end to end. And there is an important lesson from there. If we build big enough architecture and feed it enough data, we can let the automatic feature learner, deep neural networks, learn the needed nuances and complexities that we did not know existed. After all, do we know that we could have solved the problem of protein folding without deep learning?
Oskar: Thanks, Noah! Coming back to you, Hosein, and the research part your group performs at Celeris, could you talk a little more about the work you are doing there and how you manage to steer the research project in a successful direction?
Hosein: Yes, sure. As I mentioned before, one of the main challenges for applying machine learning in the targeted protein degradation domain is the sparsity of the data. As a result, one of our main focuses is developing an active learning framework to gain the most information from little data. For example, we have tried to apply active learning for ternary complex prediction (the complex formed between two proteins and PROTAC), which is a really hard task.
The other thing that we are working on is how we can use the symmetry available in our data (compounds, proteins) and design a network that respects that symmetry. We put a more inductive bias on the network (because the network should remain invariant to some transformations). We can learn something even in the small data regime by putting more inductive bias from symmetry in the data. This is where we are using the power of graph neural networks (which are permutation invariant/equivariant) or geodesic convolution in 3d point clouds.
Oskar: Makes sense, and Noah, how is the research part integrated into the production level system and eventually to the customer?
Noah: Research and the engineering part, together with (computational) chemistry and biology teams, are deeply intertwined. Once we have encoded our requirements into data and researched a novel solution, we can test, scale, and bring the software to customers. As we have discovered, these steps are not separated but are starting together with the research. Meaning we focus on the usefulness for the customers, testing the code, and scaling it as we are developing novel solutions, all while preserving the chemical and biological requirements. Meaning in conjunction with the other teams.
Oskar: Gotcha, Hosein. What about the future of Celeris from a research perspective? What vision do you have in this regard? What will be the major focus?
Hosein: Currently, biologists, chemists, and people with strong domain knowledge are mostly working in targeted protein degradation. They have done a great job and achieved some fantastic results. But I think the whole process is amenable to more rational design and less try and error. I hope we will develop some tools that can make this process faster and less error prone. I think we will be able to create tools that can help chemists design novel PROTACs or molecular glues quickly.